
みなさん、こんにちは!私の名前はボリスです。高等経済学部のデータサイエンスプログラムを卒業し、MLエンジニアとして働いており、OTUSコースでML Professional、DL Basic、ComputerVisionを教えています。
2021 “” , . : “ ”? , “” .
, . , . COVID-19 train-test split.
. , , - .
.
.
: TBA
?
COVID-19 , . () . (variants of concern). B.1.1.7, “” .
? -, , 40% - 90% , ( : R0 40% - 90% ). -, . -, , - .
, B.1.1.7 COVID-19 30 .

, . .
: 2020.03.10 - 2021.03.23.
. , , , . .
df.columns = ['date', 'region',
'total_infected', 'total_recovered', 'total_dead',
'deaths_per_day', 'infected_per_day', 'recovered_per_day']
df = df[df.region == ''].reset_index()
df['date'] = pd.to_datetime(df['date'], format='%d.%m.%Y')
df['infected'] = df['total_infected'] - df['total_recovered'] - df['total_dead']
df = df.drop(columns=['index', 'region'])
df = df.sort_values(by='date')
df.index = df.date
df['date'] = df.date.asfreq('D')
7 . .
df_smoothed = df.rolling(7).mean().round(5)
df_smoothed.columns = [col + '_ma7' for col in df_smoothed.columns]
full_df = pd.concat([df, df_smoothed], axis=1)
for column in full_df.columns:
if column.endswith('_ma7'):
original_column = column.strip('_ma7')
full_df[column] = full_df[column].fillna(full_df[original_column])

, :
infected_per_day
— .
recovered_per_day
— .
deaths_per_day
— .
total_infected
— ,infected_per_day
.
total_dead
— ,deaths_per_day
.
total_recovered
— ,recovered_per_day
.
: SEIRD
SIR , . : Susceptible, Infected, Recovered.
“ ”:
(Infected).
- Susceptible. Infected.
Infected , Recovered.
, .
SIR , SEIRD . SEIRD , :
Susceptible — .
Exposed — , , .
Infectious — .
Recovered — .
Dead — .
:

, Infected . δ E I. γ I , R D. α, (Infection Fatality Rate). , , .
:
α — case fatality rate.
β — , .
δ — 1 .
y — 1 .
R0 = β/y — , , .
SEIRD. -, SEIRD , COVID-19 . -, , , , . -, “” . , : “ , R0 90% ?” , , , .
SEIRD: . . . : , B.1.1.7 . , , , COVID-19 SEIR .
SEIRD
SEIRD.
class BarebonesSEIR:
def __init__(self, params=None):
self.params = params
def get_fit_params(self):
params = lmfit.Parameters()
params.add("population", value=12_000_000, vary=False)
params.add("epidemic_started_days_ago", value=10, vary=False)
params.add("r0", value=4, min=3, max=5, vary=True)
params.add("alpha", value=0.0064, min=0.005, max=0.0078, vary=True) # CFR
params.add("delta", value=1/3, min=1/14, max=1/2, vary=True) # E -> I rate
params.add("gamma", value=1/9, min=1/14, max=1/7, vary=True) # I -> R rate
params.add("rho", expr='gamma', vary=False) # I -> D rate
return params
def get_initial_conditions(self, data):
# Simulate such initial params as to obtain as many deaths as in data
population = self.params['population']
epidemic_started_days_ago = self.params['epidemic_started_days_ago']
t = np.arange(epidemic_started_days_ago)
(S, E, I, R, D) = self.predict(t, (population - 1, 0, 1, 0, 0))
I0 = I[-1]
E0 = E[-1]
Rec0 = R[-1]
D0 = D[-1]
S0 = S[-1]
return (S0, E0, I0, Rec0, D0)
def step(self, initial_conditions, t):
population = self.params['population']
delta = self.params['delta']
gamma = self.params['gamma']
alpha = self.params['alpha']
rho = self.params['rho']
rt = self.params['r0'].value
beta = rt * gamma
S, E, I, R, D = initial_conditions
new_exposed = beta * I * (S / population)
new_infected = delta * E
new_dead = alpha * rho * I
new_recovered = gamma * (1 - alpha) * I
dSdt = -new_exposed
dEdt = new_exposed - new_infected
dIdt = new_infected - new_recovered - new_dead
dRdt = new_recovered
dDdt = new_dead
assert S + E + I + R + D - population <= 1e10
assert dSdt + dIdt + dEdt + dRdt + dDdt <= 1e10
return dSdt, dEdt, dIdt, dRdt, dDdt
def predict(self, t_range, initial_conditions):
ret = odeint(self.step, initial_conditions, t_range)
return ret.T
.
get_fit_params
. lmfit
, , Parameters
. - COVID-19.
epidemic_started_days_ago
. 2 2020.
step
. initial_conditions
t
, .
predict
t_range
. scipy.integrate.odeint step
, t_range
.
get_initial_conditions
epidemic_started_days_ago
. .
:
model = BarebonesSEIR()
model.params = model.get_fit_params()
train_initial_conditions = model.get_initial_conditions(train_subset)
train_t = np.arange(len(train_subset))
(S, E, I, R, D) = model.predict(train_t, train_initial_conditions)
plt.figure(figsize=(10, 7))
plt.plot(train_subset.date, train_subset['total_dead'], label='ground truth')
plt.plot(train_subset.date, D, label='predicted', color='black', linestyle='dashed' )
plt.legend()
plt.title('Total deaths')
plt.show()

, , . , .
SEIRD:

“” , , . , “” . , .
“” . , t, R0 q(t). : Rt = R0 - R0 * q(t). β(t) = Rt * y .
q(t). . 60 , : . , .
.
def sigmoid(x, xmin, xmax, a, b, c, r):
x_scaled = (x - xmin) / (xmax - xmin)
out = (a * np.exp(c * r) + b * np.exp(r * x_scaled)) / (np.exp(c * r) + np.exp(x_scaled * r))
return out
def stepwise_soft(t, coefficients, r=20, c=0.5):
t_arr = np.array(list(coefficients.keys()))
min_index = np.min(t_arr)
max_index = np.max(t_arr)
if t <= min_index:
return coefficients[min_index]
elif t >= max_index:
return coefficients[max_index]
else:
index = np.min(t_arr[t_arr >= t])
if len(t_arr[t_arr < index]) == 0:
return coefficients[index]
prev_index = np.max(t_arr[t_arr < index])
# sigmoid smoothing
q0, q1 = coefficients[prev_index], coefficients[index]
out = sigmoid(t, prev_index, index, q0, q1, c, r)
return out
t_range = np.arange(100)
coefficients = {
0: 0,
30: 0.5,
60: 1,
100: 0.4,
}
plt.title(' ')
plt.scatter(coefficients.keys(), coefficients.values(), label=' ')
plt.plot(t_range, [stepwise_soft(t, coefficients, r=20, c=0.5) for t in t_range], label=' ')
plt.xlabel('t')
plt.ylabel(' ')
plt.legend(loc='lower center', bbox_to_anchor=(0.5, -0.6),)
plt.show()

SEIRD :
get_fit_params
. , . , :stepwise_size
, 60 .
def get_fit_params(self, data):
...
params.add(f"t0_q", value=0, min=0, max=0.99, brute_step=0.1, vary=False)
piece_size = self.stepwise_size
for t in range(piece_size, len(data), piece_size):
params.add(f"t{t}_q", value=0.5, min=0, max=0.99, brute_step=0.1, vary=True)
return params
get_step_rt_beta
, β(t) Rt .
step
get_step_rt_beta
, β(t)t
.
, . , . .
Infected, Recovered Dead :
Iv(t) — , .
I(t) — , .
Rv(t) — , .
R(t) — , .
Dv(t) — , .
D(t) — , .
:
pi — , Iv.
pd — , , Dv. Iv Dv, , COVID-19.
, , :

SEIRD . SEIRD . .
, , Iv(t), Rv(t) Dv(t) . .
, lmfit.minimize. callable, (, residuals), , . Levenberg-Marquardt (method=’leastsq’
), , . , , , , stepwise_size
.
BaseFitter
, . minimize
.
:
def smape_resid_transform(true, pred, eps=1e-5):
return (true - pred) / (np.abs(true) + np.abs(pred) + eps)
class HiddenCurveFitter(BaseFitter):
...
def residual(self, params, t_vals, data, model):
model.params = params
initial_conditions = model.get_initial_conditions(data)
(S, E, I, Iv, R, Rv, D, Dv), history = model.predict(t_vals, initial_conditions, history=False)
(new_exposed,
new_infected_invisible, new_infected_visible,
new_recovered_invisible,
new_recovered_visible,
new_dead_invisible, new_dead_visible) = model.compute_daily_values(S, E, I, Iv, R, Rv, D, Dv)
new_infected_visible = new_infected_visible
new_dead_visible = new_dead_visible
new_recovered_visible = new_recovered_visible
true_daily_cases = data[self.new_cases_col].values[1:]
true_daily_deaths = data[self.new_deaths_col].values[1:]
true_daily_recoveries = data[self.new_recoveries_col].values[1:]
resid_I_new = smape_resid_transform(true_daily_cases, new_infected_visible)
resid_D_new = smape_resid_transform(true_daily_deaths, new_dead_visible)
resid_R_new = smape_resid_transform(true_daily_recoveries, new_recovered_visible)
if self.weights:
residuals = np.concatenate([
self.weights['I'] * resid_I_new,
self.weights['D'] * resid_D_new,
self.weights['R'] * resid_R_new,
]).flatten()
else:
residuals = np.concatenate([
resid_I_new,
resid_D_new,
resid_R_new,
]).flatten()
return residuals
. t_vals
, . .
, .
-, : 1000 1 . , “ ” . smape_resid_transform
.
-, , . (self.weights
) . : 0.5
0.25
.
, SEIRD . . .
from sir_models.fitters import HiddenCurveFitter
from sir_models.models import SEIRHidden
stepwize_size = 60
weights = {
'I': 0.25,
'R': 0.25,
'D': 0.5,
}
model = SEIRHidden(stepwise_size=stepwize_size)
fitter = HiddenCurveFitter(
new_deaths_col='deaths_per_day_ma7',
new_cases_col='infected_per_day_ma7',
new_recoveries_col='recovered_per_day_ma7',
weights=weights,
max_iters=1000,
)
fitter.fit(model, train_subset)
result = fitter.result
result
:

:

! , . : .
: , . , 10 , .

, . , : . . , “ ”, .
c -
, . , , — .
COVID-19 , .
:
, 20- .
:
.
30 .
.
.
: .
from sir_models.utils import eval_on_select_dates_and_k_days_ahead
from sir_models.utils import smape
from sklearn.metrics import mean_absolute_error
K = 30
last_day = train_subset.date.iloc[-1] - pd.to_timedelta(K, unit='D')
eval_dates = pd.date_range(start='2020-06-01', end=last_day)[::20]
def eval_hidden_moscow(train_df, t, train_t, eval_t):
weights = {
'I': 0.25,
'R': 0.25,
'D': 0.5,
}
model = SEIRHidden()
fitter = HiddenCurveFitter(
new_deaths_col='deaths_per_day_ma7',
new_cases_col='infected_per_day_ma7',
new_recoveries_col='recovered_per_day_ma7',
weights=weights,
max_iters=1000,
save_params_every=500)
fitter.fit(model, train_df)
train_initial_conditions = model.get_initial_conditions(train_df)
train_states, history = model.predict(train_t, train_initial_conditions, history=False)
test_initial_conds = [compartment[-1] for compartment in train_states]
test_states, history = model.predict(eval_t, test_initial_conds, history=False)
return model, fitter, test_states
(models, fitters,
model_predictions,
train_dfs, test_dfs) = eval_on_select_dates_and_k_days_ahead(train_subset,
eval_func=eval_hidden_moscow,
eval_dates=eval_dates,
k=K)
model_pred_D = [pred[7] for pred in model_predictions]
true_D = [tdf.total_dead.values for tdf in test_dfs]
baseline_pred_D = [[tdf.iloc[-1].total_dead]*K for tdf in train_dfs]
overall_errors_model = [mean_absolute_error(true, pred) for true, pred in zip(true_D, model_pred_D)]
overall_errors_baseline = [mean_absolute_error(true, pred) for true, pred in zip(true_D, baseline_pred_D)]
print('Mean overall error baseline', np.mean(overall_errors_baseline).round(3))
print('Mean overall error model', np.mean(overall_errors_model).round(3))
overall_smape_model = [smape(true, pred) for true, pred in zip(true_D, model_pred_D)]
np.median(overall_smape_model)

:
Mean Absolute Arror : 714.
Mean Absolute Arror : 550.
Symmetric Mean Absolute Percentage Error: 4.6%
5% , .

, . . SEIR , , I(t) : I1(t) I2(t).
SARS-CoV-2 . “” . B.1.1.7 R0: 1.4 - 1.9 . SARS-CoV-2, R0 B.1.1.7.
:

, , . beta2_mult
, , β1(t) , β2(t) .
class SEIRHiddenTwoStrains(SEIRHidden):
...
@classmethod
def from_strain_one_model(cls, model):
strain1_params = model.params
strain1_params.add("beta2_mult", value=1.5, min=1, max=2, vary=False)
return cls(params=deepcopy(strain1_params))
, , , , .
B.1.1.7
, . R0 B.1.1.7, . .
R0 . B.1.1.7 -. .
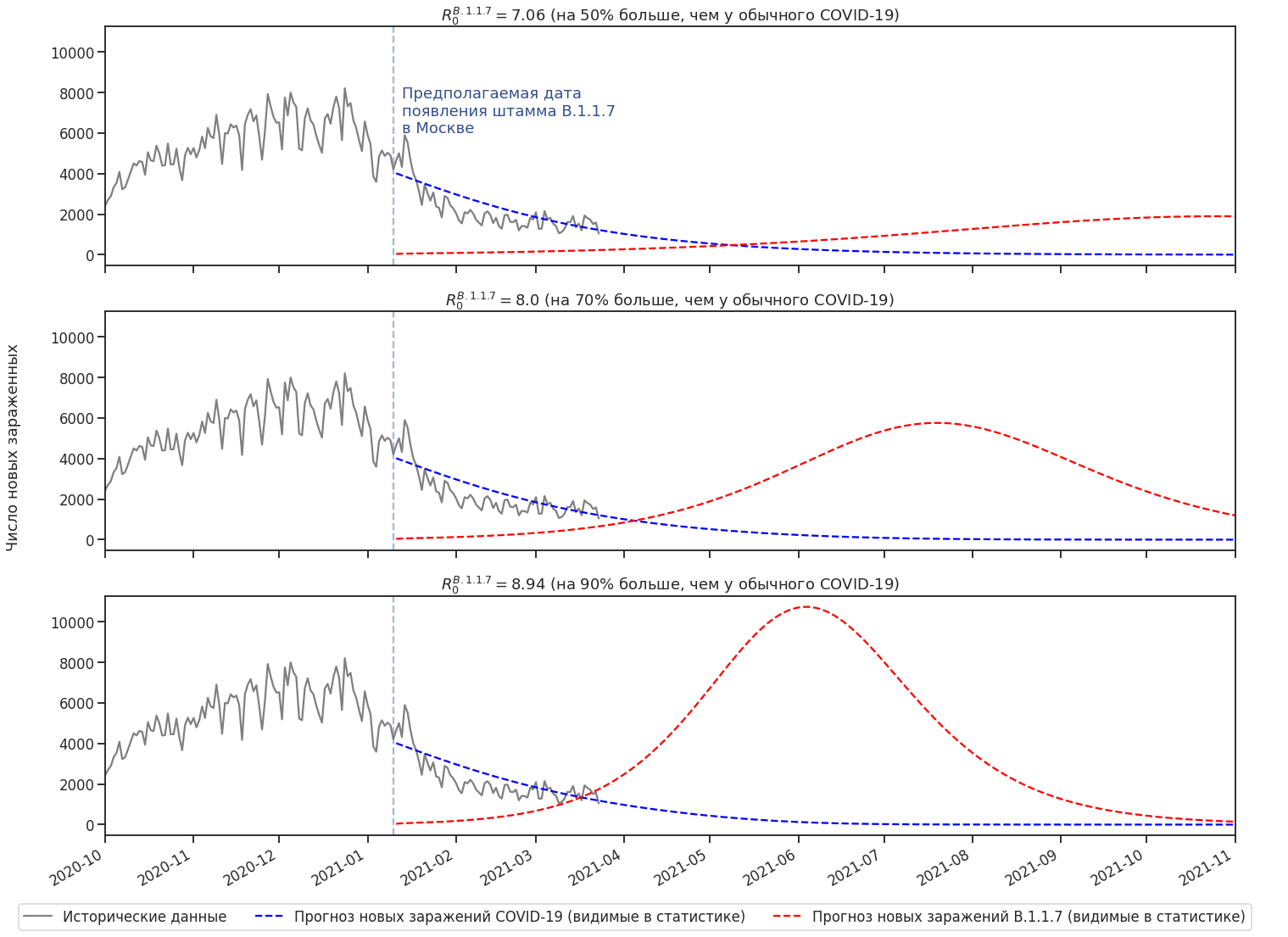
“ ” . .

.

:
"Machine Learning. Professional".
: « ML : ».